By: Gareth Willmer
Send to a friend
The details you provide on this page will not be used to send unsolicited email, and will not be sold to a 3rd party. See privacy policy.
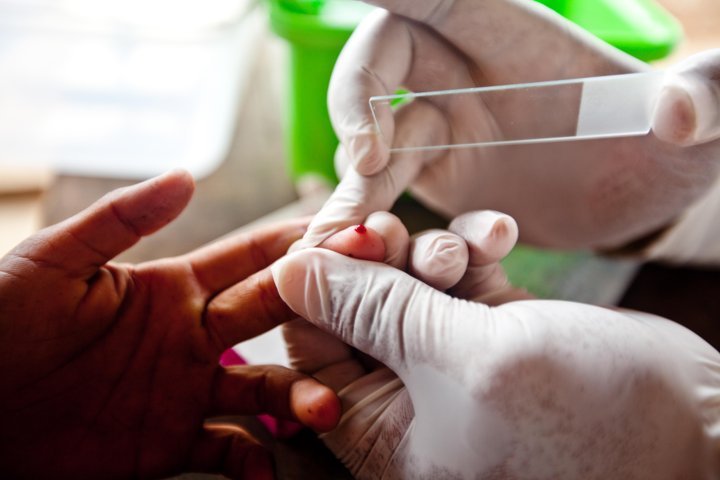
A self-styled ‘ultrasensitive’ malaria test could lead to more accurate identification of the potentially significant pool of people who carry the disease without showing any symptoms, a paper has found.
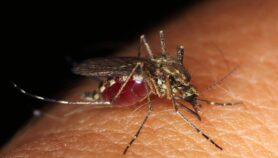
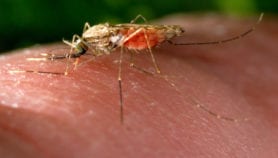
The paper indicates that these ‘asymptomatic carriers’ are far more common than thought. They are an emerging front line in the fight to eradicate malaria, as they act as a hidden reservoir, allowing the parasite to carry on being circulated from humans to mosquitoes.
In the past, malaria was detected by using a light microscope to spot parasites in blood samples. But microbiological tests that pick up the parasites’ genetic signature indicate that, in areas with low levels of malaria, asymptomatic carriers may account for a fifth to a half of all parasites passed from humans to mosquitoes.
“The main significance of our findings is that we have identified a hidden parasite population that cannot be detected even using standard molecular methods.”
Ingrid Felger, Tropical and Public Health Institute
Ingrid Felger of the Swiss Tropical and Public Health Institute in Switzerland, whose team worked on the new test, says asymptomatic carriers may account for as much as 70 per cent of malaria infections in some locations.
Felger tells SciDev.Net that scientists think asymptomatic carriers are most prevalent in places that have seen a rapid drop in infection levels, either naturally or due to eradication programmes. These include island populations such as Zanzibar in Tanzania, where her team carried out its study.
This pattern of prevalence means the problem is likely to grow as malaria control improves, Felger says.
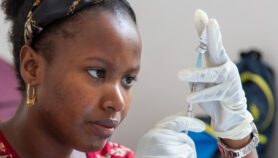
In a paper published last month in PLOS Medicine, the researchers built on existing biomolecular methods for malaria detection that use a technique called quantitative polymerase chain reaction (qPCR). This involves amplifying DNA from human blood to find parasite-specific sequences.
But Felger’s method targets sequences of parasite DNA that occur between 60 and 250 times in the parasite’s genetic code, as opposed to standard tests, which target sequences that occur just five times. This should help cut the number of “false negatives” test produce, she says.
The team found that the new test was about ten times more sensitive than existing ones. In addition, the authors found that when standard molecular-diagnostics tests were used, the presence of malaria parasites was underestimated by 8 per cent compared with the new tests.
“The main significance of our findings is that we have identified a hidden parasite population that cannot be detected even using standard molecular methods,” says Felger. “This teaches us that asymptomatic, low-density malaria infections are much more prevalent than anticipated.”
But the test has limitations. For instance, it requires advanced laboratory equipment. And it only targets the main malarial killer, Plasmodium falciparum, not other Plasmodium species that cause the disease.
Andrea Bosman, who coordinates the diagnosis, treatment and vaccines unit in the WHO’s Global Malaria Programme, says the new test is an improvement on conventional qPCR methods in terms of aiding detection.
Bosman thinks the paper is not a “major breakthrough”. But he points out that other researchers are investigating malaria detection using another type of biological marker: antigens, the unique proteins found on the parasites’ cells.
“This is a big area of research with increasing attention,” says Bosman. “We only see the tip of the iceberg at the moment.”
References
Natalie Hofmann and others Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets (PLOS Medicine, 3 March 2015)